Structural Insight into Evolution of the Quinone Binding Site in Complex II

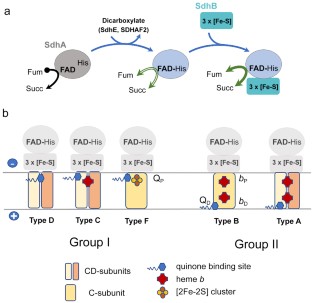
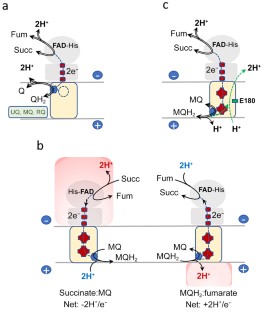
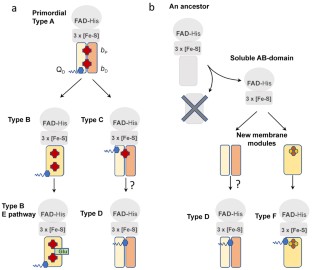
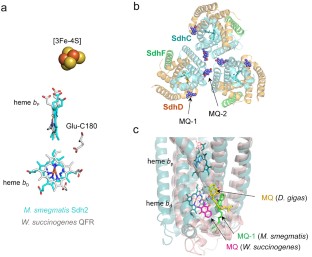
The Complex II family encompasses membrane bound succinate:quinones reductases and quinol:fumarate reductases that catalyze interconversion of succinate and fumarate coupled with reduction and oxidation of quinone. These enzymes are found in all biological genres and share a modular structure where a highly conserved soluble domain is bound to a membrane-spanning domain that is represented by distinct variations. The current classification of the complex II family members is based on the number of subunits and co-factors in the membrane anchor (types A-F). This classification also provides insights into possible evolutionary paths and suggests that some of the complex II enzymes (types A-C) co-evolved as the whole assembly. Origin of complex II types D and F may have arisen from independent events of de novo association of the conserved soluble domain with a new anchor. Here we analyze a recent structure of Mycobacterium smegmatis Sdh2, a complex II enzyme with two transmembrane subunits and two heme b molecules. This analysis supports an earlier hypothesis suggesting that mitochondrial complex II (type C) with a single heme b may have evolved as an assembled unit from an ancestor similar to M. smegmatis Sdh2.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Subscribe and save
Springer+ Basic
€32.70 /Month
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
Buy Now
Price includes VAT (France)
Instant access to the full article PDF.
Rent this article via DeepDyve
Similar content being viewed by others

Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF
Article Open access 25 August 2020

Rieske/Cytochrome b Complexes: The Turbo Chargers of Chemiosmosis
Chapter © 2014

Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis
Article 05 December 2018
Abbreviations
distal heme b
proximal heme b
distal quinone binding site
proximal quinone binding site
References
- Cecchini, G. (2003) Function and structure of complex II of the respiratory chain, Annu. Rev. Biochem., 72, 77-109, https://doi.org/10.1146/annurev.biochem.72.121801.161700. ArticleCASPubMedGoogle Scholar
- Hagerhall, C. (1997) Succinate: quinone oxidoreductases. Variations on a conserved theme, Biochim. Biophys. Acta, 1320, 107-141, https://doi.org/10.1016/s0005-2728(97)00019-4. ArticleCASPubMedGoogle Scholar
- Sharma, P., Maklashina, E., Cecchini, G., and Iverson, T. M. (2019) Maturation of the respiratory complex II flavoprotein, Curr. Opin. Struct. Biol., 59, 38-46, https://doi.org/10.1016/j.sbi.2019.01.027. ArticleCASPubMedPubMed CentralGoogle Scholar
- Sharma, P., Maklashina, E., Cecchini, G., and Iverson, T. M. (2018) Crystal structure of an assembly intermediate of respiratory Complex II, Nat. Commun., 9, 274, https://doi.org/10.1038/s41467-017-02713-8. ArticleCASPubMedPubMed CentralGoogle Scholar
- Kounosu, A. (2014) Analysis of covalent flavinylation using thermostable succinate dehydrogenase from Thermus thermophilus and Sulfolobus tokodaii lacking SdhE homologs, FEBS Lett., 588, 1058-1063, https://doi.org/10.1016/j.febslet.2014.02.022. ArticleCASPubMedGoogle Scholar
- Lill, R., and Freibert, S. A. (2020) Mechanisms of mitochondrial iron-sulfur protein biogenesis, Annu. Rev. Biochem., 89, 471-499, https://doi.org/10.1146/annurev-biochem-013118-111540. ArticleCASPubMedGoogle Scholar
- Bai, Y., Chen, T., Happe, T., Lu, Y., and Sawyer, A. (2018) Iron-sulphur cluster biogenesis via the SUF pathway, Metallomics, 10, 1038-1052, https://doi.org/10.1039/c8mt00150b. ArticlePubMedGoogle Scholar
- Heuts, D. P., Scrutton, N. S., McIntire, W. S., and Fraaije, M. W. (2009) What’s in a covalent bond? On the role and formation of covalently bound flavin cofactors, FEBS J., 276, 3405-3427, https://doi.org/10.1111/j.1742-4658.2009.07053.x. ArticleCASPubMedGoogle Scholar
- Hao, H. X., Khalimonchuk, O., Schraders, M., Dephoure, N., Bayley, J. P., et al. (2009) SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma, Science, 325, 1139-1142, https://doi.org/10.1126/science.1175689. ArticleCASPubMedGoogle Scholar
- McNeil, M. B., Clulow, J. S., Wilf, N. M., Salmond, G. P., and Fineran, P. C. (2012) SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria, J. Biol. Chem., 287, 18418-18428, https://doi.org/10.1074/jbc.M111.293803. ArticleCASPubMedPubMed CentralGoogle Scholar
- Van Vranken, J. G., Na, U., Winge, D. R., and Rutter, J. (2015) Protein-mediated assembly of succinate dehydrogenase and its cofactors, Crit. Rev. Biochem. Mol. Biol., 50, 168-180, https://doi.org/10.3109/10409238.2014.990556. ArticleCASPubMedGoogle Scholar
- Moosavi, B., Berry, E. A., Zhu, X. L., Yang, W. C., and Yang, G. F. (2019) The assembly of succinate dehydrogenase: a key enzyme in bioenergetics, Cell. Mol. Life Sci., 76, 4023-4042, https://doi.org/10.1007/s00018-019-03200-7. ArticleCASPubMedGoogle Scholar
- Tedeschi, G., Negri, A., Mortarino, M., Ceciliani, F., Simonic, T., et al. (1996) L-aspartate oxidase from Escherichia coli. II. Interaction with C4 dicarboxylic acids and identification of a novel L-aspartate: fumarate oxidoreductase activity, Eur. J. Biochem., 239, 427-433, https://doi.org/10.1111/j.1432-1033.1996.0427u.x. ArticleCASPubMedGoogle Scholar
- Taylor, P., Pealing, S. L., Reid, G. A., Chapman, S. K., and Walkinshaw, M. D. (1999) Structural and mechanistic mapping of a unique fumarate reductase, Nat. Struct. Biol., 6, 1108-1112, https://doi.org/10.1038/70045. ArticleCASPubMedGoogle Scholar
- Maklashina, E., Rajagukguk, S., Iverson, T. M., and Cecchini, G. (2018) The unassembled flavoprotein subunits of human and bacterial complex II have impaired catalytic activity and generate only minor amounts of ROS, J. Biol. Chem., 293, 7754-7765, https://doi.org/10.1074/jbc.RA118.001977. ArticleCASPubMedPubMed CentralGoogle Scholar
- Maklashina, E., Iverson, T. M., Sher, Y., Kotlyar, V., Andrell, J., et al. (2006) Fumarate reductase and succinate oxidase activity of Escherichia coli complex II homologs are perturbed differently by mutation of the flavin binding domain, J. Biol. Chem., 281, 11357-11365, https://doi.org/10.1074/jbc.M512544200. ArticleCASPubMedGoogle Scholar
- Hägerhäll, C., and Hederstedt, L. (1996) A structural model for the membrane-integral domain of succinate: quinone oxidoreductases, FEBS Lett., 389, 25-31, https://doi.org/10.1016/0014-5793(96)00529-7. ArticlePubMedGoogle Scholar
- Lancaster, C. R. (2013) The di-heme family of respiratory complex II enzymes, Biochim. Biophys. Acta, 1827, 679-687, https://doi.org/10.1016/j.bbabio.2013.02.012. ArticleCASPubMedGoogle Scholar
- Iverson, T. M., Luna-Chavez, C., Cecchini, G., and Rees, D. C. (1999) Structure of the Escherichia coli fumarate reductase respiratory complex, Science, 284, 1961-1966, https://doi.org/10.1126/science.284.5422.1961. ArticleCASPubMedGoogle Scholar
- Yankovskaya, V., Horsefield, R., Tornroth, S., Luna-Chavez, C., Miyoshi, H., et al. (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation, Science, 299, 700-704, https://doi.org/10.1126/science.1079605. ArticleCASPubMedGoogle Scholar
- Huang, L. S., Shen, J. T., Wang, A. C., and Berry, E. A. (2006) Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the “oxaloacetate-inhibited” state, Biochim. Biophys. Acta, 1757, 1073-1083, https://doi.org/10.1016/j.bbabio.2006.06.015. ArticleCASPubMedPubMed CentralGoogle Scholar
- Sun, F., Huo, X., Zhai, Y., Wang, A., Xu, J., et al. (2005) Crystal structure of mitochondrial respiratory membrane protein complex II, Cell, 121, 1043-1057, https://doi.org/10.1016/j.cell.2005.05.025. ArticleCASPubMedGoogle Scholar
- Inaoka, D. K., Shiba, T., Sato, D., Balogun, E. O., Sasaki, T., et al. (2015) Structural insights into the molecular design of flutolanil derivatives targeted for fumarate respiration of parasite mitochondria, Int. J. Mol. Sci., 16, 15287-15308, https://doi.org/10.3390/ijms160715287. ArticleCASPubMedPubMed CentralGoogle Scholar
- Hards, K., Rodriguez, S. M., Cairns, C., and Cook, G. M. (2019) Alternate quinone coupling in a new class of succinate dehydrogenase may potentiate mycobacterial respiratory control, FEBS Lett., 593, 475-486, https://doi.org/10.1002/1873-3468.13330. ArticleCASPubMedGoogle Scholar
- Zhou, X., Gao, Y., Wang, W., Yang, X., Yang, X., et al. (2021) Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster, Proc. Natl. Acad. Sci. USA, 118, e2022308118, https://doi.org/10.1073/pnas.2022308118. ArticleCASPubMedPubMed CentralGoogle Scholar
- Lancaster, C. R., Kroger, A., Auer, M., and Michel, H. (1999) Structure of fumarate reductase from Wolinella succinogenes at 2.2 Å resolution, Nature, 402, 377-385, https://doi.org/10.1038/46483. ArticleCASPubMedGoogle Scholar
- Guan, H. H., Hsieh, Y. C., Lin, P. J., Huang, Y. C., Yoshimura, M., et al. (2018) Structural insights into the electron/proton transfer pathways in the quinol:fumarate reductase from Desulfovibrio gigas, Sci. Rep., 8, 14935, https://doi.org/10.1038/s41598-018-33193-5. ArticleCASPubMedPubMed CentralGoogle Scholar
- Gong, H., Gao, Y., Zhou, X., Xiao, Y., Wang, W., et al. (2020) Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF, Nat. Commun., 11, 4245, https://doi.org/10.1038/s41467-020-18011-9. ArticleCASPubMedPubMed CentralGoogle Scholar
- Juhnke, H. D., Hiltscher, H., Nasiri, H. R., Schwalbe, H., and Lancaster, C. R. (2009) Production, characterization and determination of the real catalytic properties of the putative “succinate dehydrogenase” from Wolinella succinogenes, Mol. Microbiol., 71, 1088-1101, https://doi.org/10.1111/j.1365-2958.2008.06581.x. ArticleCASPubMedPubMed CentralGoogle Scholar
- Tran, Q. M., Rothery, R. A., Maklashina, E., Cecchini, G., and Weiner, J. H. (2007) Escherichia coli succinate dehydrogenase variant lacking the heme b, Proc. Natl. Acad. Sci. USA, 104, 18007-18012, https://doi.org/10.1073/pnas.0707732104. ArticlePubMedPubMed CentralGoogle Scholar
- Maklashina, E., Hellwig, P., Rothery, R. A., Kotlyar, V., Sher, Y., et al. (2006) Differences in protonation of ubiquinone and menaquinone in fumarate reductase from Escherichia coli, J. Biol. Chem., 281, 26655-26664, https://doi.org/10.1074/jbc.M602938200. ArticlePubMedGoogle Scholar
- Schirawski, J., and Unden, G. (1998) Menaquinone-dependent succinate dehydrogenase of bacteria catalyzes reversed electron transport driven by the proton potential, Eur. J. Biochem, 257, 210-215, https://doi.org/10.1046/j.1432-1327.1998.2570210.x. ArticleCASPubMedGoogle Scholar
- Madej, M. G., Nasiri, H. R., Hilgendorff, N. S., Schwalbe, H., Unden, G., et al. (2006) Experimental evidence for proton motive force-dependent catalysis by the diheme-containing succinate:menaquinone oxidoreductase from the Gram-positive bacterium Bacillus licheniformis, Biochemistry, 45, 15049-15055, https://doi.org/10.1021/bi0618161. ArticleCASPubMedGoogle Scholar
- Madej, M. G., Nasiri, H. R., Hilgendorff, N. S., Schwalbe, H., and Lancaster, C. R. (2006) Evidence for transmembrane proton transfer in a dihaem-containing membrane protein complex, EMBO J., 25, 4963-4970, https://doi.org/10.1038/sj.emboj.7601361. ArticleCASPubMedPubMed CentralGoogle Scholar
- Schafer, G., Anemuller, S., and Moll, R. (2002) Archaeal complex II: “classical” and “non-classical” succinate:quinone reductases with unusual features, Biochim. Biophys. Acta, 1553, 57-73, https://doi.org/10.1016/s0005-2728(01)00232-8. ArticleCASPubMedGoogle Scholar
- Maklashina, E., Rajagukguk, S., McIntire, W. S., and Cecchini, G. (2010) Mutation of the heme axial ligand of Escherichia coli succinate-quinone reductase: implications for heme ligation in mitochondrial complex II from yeast, Biochim. Biophys. Acta, 1797, 747-754, https://doi.org/10.1016/j.bbabio.2010.01.019. ArticleCASPubMedPubMed CentralGoogle Scholar
- Lancaster, C. R., Gorss, R., Haas, A., Ritter, M., Mantele, W., et al. (2000) Essential role of Glu-C66 for menaquinol oxidation indicates transmembrane electrochemical potential generation by Wolinella succinogenes fumarate reductase, Proc. Natl. Acad. Sci. USA, 97, 13051-13056, https://doi.org/10.1073/pnas.220425797. ArticleCASPubMedPubMed CentralGoogle Scholar
- Maklashina, E., and Cecchini, G. (2010) The quinone-binding and catalytic site of complex II, Biochim. Biophys. Acta, 1797, 1877-1882, https://doi.org/10.1016/j.bbabio.2010.02.015. ArticleCASPubMedPubMed CentralGoogle Scholar
- Silkin, Y., Oyedotun, K. S., and Lemire, B. D. (2007) The role of Sdh4p Tyr-89 in ubiquinone reduction by the Saccharomyces cerevisiae succinate dehydrogenase, Biochim. Biophys. Acta, 1767, 143-150, https://doi.org/10.1016/j.bbabio.2006.11.017. ArticleCASPubMedGoogle Scholar
- Tran, Q. M., Rothery, R. A., Maklashina, E., Cecchini, G., and Weiner, J. H. (2006) The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b, J. Biol. Chem., 281, 32310-32317, https://doi.org/10.1074/jbc.M607476200. ArticleCASPubMedGoogle Scholar
- Tran, Q. M., Fong, C., Rothery, R. A., Maklashina, E., Cecchini, G., and Weiner, J. H. (2012) Out of plane distortions of the heme b of Escherichia coli succinate dehydrogenase, PLoS One, 7, e32641, https://doi.org/10.1371/journal.pone.0032641. ArticleCASPubMedPubMed CentralGoogle Scholar
- Maklashina, E., Rothery, R. A., Weiner, J. H., and Cecchini, G. (2001) Retention of heme in axial ligand mutants of succinate-ubiquinone oxidoreductase (complex II) from Escherichia coli, J. Biol. Chem., 276, 18968-18976, https://doi.org/10.1074/jbc.M011270200. ArticleCASPubMedGoogle Scholar
- Gu, L. Q., Yu, L., and Yu, C. A. (1990) Effect of substituents of the benzoquinone ring on electron-transfer activities of ubiquinone derivatives, Biochim. Biophys. Acta, 1015, 482-492, https://doi.org/10.1016/0005-2728(90)90082-f. ArticleCASPubMedGoogle Scholar
Acknowledgments
I dedicate this review to my mentor Andrei Vinogradov. He devoted a large portion of his scientific life to study mitochondrial complex II, which he often called “my first love in science”. I also wish to thank Gary Cecchini for reading the manuscript.
Author information
Authors and Affiliations
- Department of Biochemistry and Biophysics, University of California, San Francisco, 94143, San Francisco, CA, USA Elena Maklashina
- Elena Maklashina








